Products Description
適用於Illumina測序儀,高通量優化,dual-indexed DNA片段建庫製備
- 高通量格式:提供足夠的試劑,以實現自動或手動多通道移液管96孔盤library建構
- 易於使用:兩色雙索引(index)套組有助於觀察primer正確的添加
- 與NxSeq®UltraLowDNA Library Kit相同的性能特徵,包括:
- High Quality Data: 極高的adaptor連接效率,可得到 sequencing depth 一致性及絕佳的基因覆蓋率
- Sensitive: 檢體量兼容範圍寬50 pg~ 75 ng of sheared/fragmented DNA
- Minimal Bias: 有力,統一的PCR擴增, 以提高基因覆蓋率的一致性
- Flexible: 可用於de novo whole genome sequencing, resequencing, exome-seq, ChIP-seq and FFPE DNA檢體
- 高價值:高經濟效益的建庫和索引(index)套組以產生出色的測序結果
Dual Index Library Construction
|
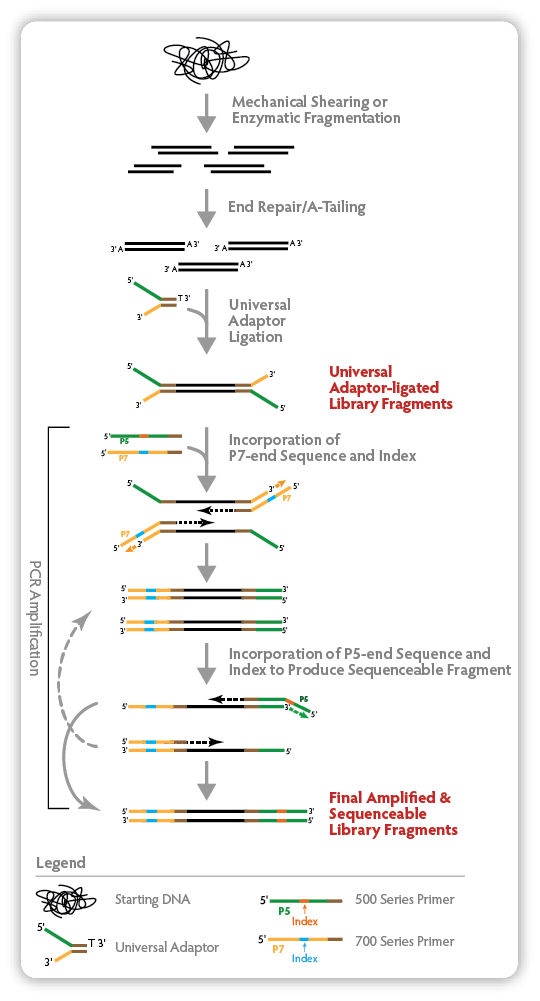
|
| Figure 1. Mechanics of NxSeq® UltraLow Kit library construction using dual indexing primers. This figure illustrates how DNA fragment libraries are constructed using the NxSeq® UltraLow Library Prep Kit and the NxSeq® HT Dual Indexing Kit. Each final library fragment has a 500 Series Index at one end and a 700 Series Index at the other end. Note that some PCR fragments and products are not shown to simplify the illustration. |
Colored Dual Indexing Primer Sets Provide a Visual Verification of Correct Dispensing
|
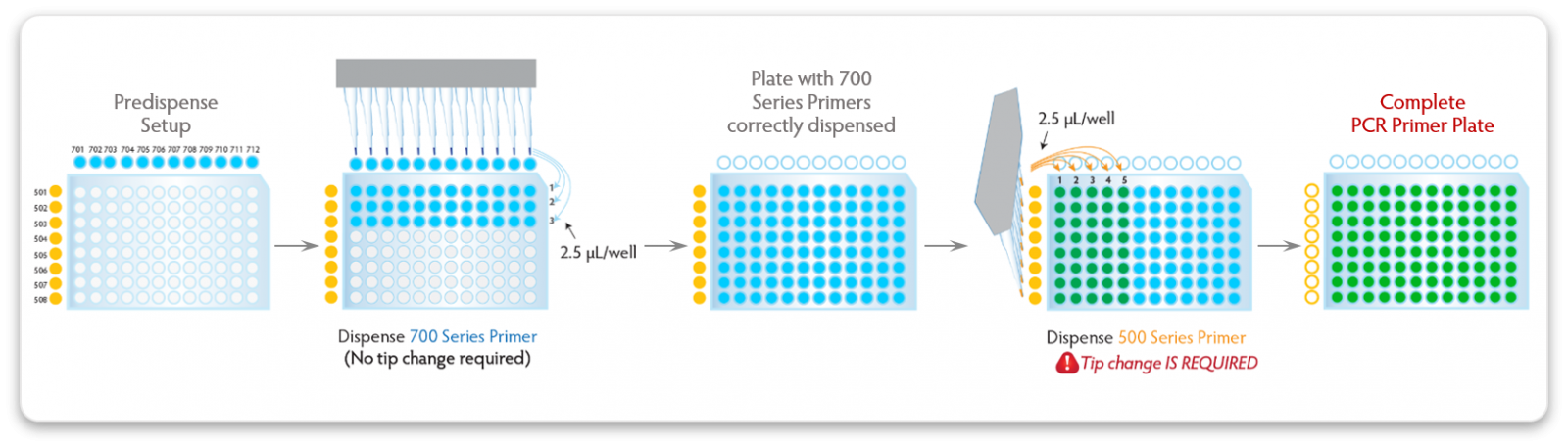
|
| Figure 2. 96-well PCR plate dual index primer dispensing protocol. The 700 Series Indexing Primers contain an inert blue dye and the 500 Series Indexing Primers contain an inert yellow/orange dye. The 700 Series Primers are dispensed first, row by row. As primers are dispensed, the rows become blue. The 500 Series Primers are then dispensed column by column, and as they are added the well color changes from blue to green (blue + yellow = green). After dispensing is complete, all wells should exhibit a consistent green color. The presence of clear, yellow/orange or blue wells indicates that those wells did not get the appropriate 700 and/or 500 Series Primers. |
The Inert Dyes Added to the Dual Indexing Primers have No Effect on Sequencing Results
|
| |
Triplicate Human Libraries |
| |
(+) Dye |
(-) Dye |
| Average Number of Reads |
145,192,042 |
149,394,251 |
| Percent Mapped Reads |
95.4% |
95.4% |
| Insert Length |
258 bp |
255 bp |
| Average Depth |
5.3 |
5.3 |
| Proper Pair Duplication Rate |
8.4% |
8.8% |
| Autosome Coverage |
96.4% |
95.8% |
|
Figure 3. Analyzing the effect of the inert dyes on sequencing results. Sheared human gDNA was generated by mechanical shearing (Covaris LE220, peak size 300 bp) and six libraries were constructed using 10 ng of sheared gDNA input per library and the NxSeq® UltraLow DNA Library Kit, 96 Reactions according to the User Manual up through Universal Adaptor Ligation and Bead Cleanup steps. After cleanup, the 6 adaptor-ligated libraries were pooled, mixed and split into six PCR reactions. The two inert dyes present in the NxSeq® HT Dual Indexing Primers were then added to three PCR reactions with each dye at the same concentration as in PCR reactions set up using the NxSeq HT Dual Indexing Primers. An equivalent volume of water was added to the other three PCR reactions. Indexing primers were added to each reaction (different indices) followed by PCR amplification of all libraries, library Cleanup and Size Selection. The final libraries were sent to Hudson Alpha for sequencing on the HiSeq X Ten. Note that ExAmp duplicates caused by the instrument were not removed bioinformatically. The results presented are the average results for each set of triplicate libraries. |
Consistent Library Yields Across a 96-well Plate
|
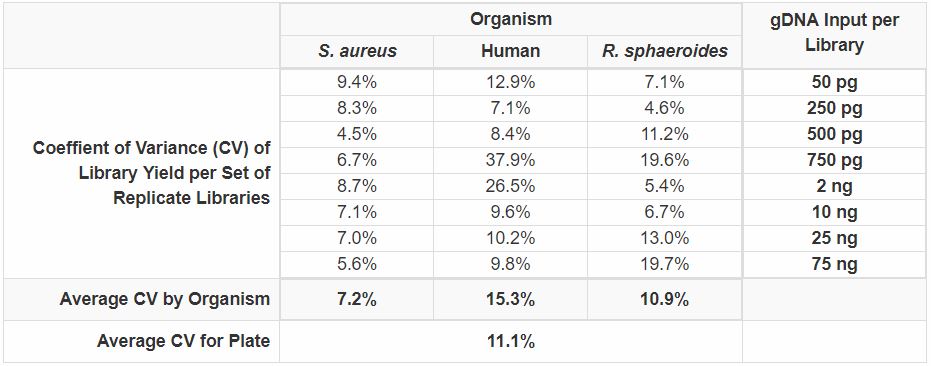
|
| Figure 4. Analysis of variation in library yield for libraries generated in a 96-well plate using the multichannel pipette protocol.Sheared gDNA was generated by mechanical shearing (Covaris LE220, peak size 300 bp) for the three genomes indicated. The indicated amounts of each gDNA were dispensed into a 96-well plate with four replicates of each input amount per organism and libraries were constructed using the NxSeq® UltraLow DNA Library Kit, 96 Reactions and the NxSeq® HT Dual Indexing Kit according to the User Manual (MA169). Note that the libraries had to be removed from the plate for PCR amplification due to the different number of PCR cycles per gDNA input amount, but all other steps including AMPure XP bead cleanup and size selection steps were performed in a single 96-well plate. Final libraries were quantitated on a Qubit® Fluorometer and the coefficient of variation (CV %) for each set of replicates was determined. The average CV% by organism (columns) and across the entire plate (last row) were also determined. Note that there were four outliers that significantly increased the already low plate CV and by removing them from the analysis the CV drops to 8%. |
High Quality Deep-sequencing Results from Human Libraries Made in 96-well Plates
|
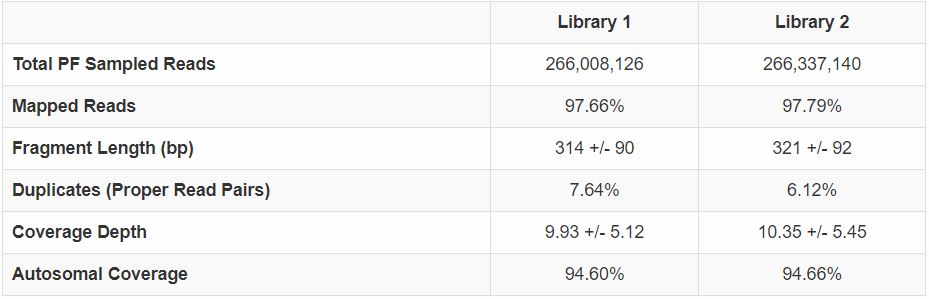 |
Figure 5. HiSeq 2500 sequencing of two human libraries prepared in a 96-well plate using the multi-channel pipette protocol. Two of the human 2 ng libraries prepped in the Figure 3 experiment were sequenced on a HiSeq 2500 using 2 x 150 bp chemistry and analyzed. |
.png) Manual下載
Manual下載
Information
The NxSeq® UltraLow DNA Library Kit, 96 Reactions and NxSeq® HT Dual Indexing Kit are only compatible with Illumina sequencers.
The NxSeq® UltraLow DNA Library Kit, 96 Reactions contains Enzyme Mix (EM), 2X Buffer (2XB), Ligase (LIG), 2X PCR Master Mix (MM) and Elution Buffer (EB). A NxSeq® HT Dual Indexing Kit is required to complete library prep and must be purchased separately.
The NxSeq® HT Dual Indexing Kit contains a Universal Adaptor, Adaptor Dilution Buffer and Indexing Primers 701 – 712, and 501 – 508. There is enough of each reagent in this kit to generate 96 different dual indexed libraries. Each NxSeq® HT Dual Index equals the TruSeq® HT Index with the same number.
Resources